by Matt Hall
Why do you like Star Trek? I’ll bet if I threw that one out there I’d get a huge variety of reasons, since there’s a lot to like.
That said, those answers tend to fall into two different categories, from “its progressive, optimistic vision of the future” to “it’s a future where a huge variety of species come together to make the galaxy a better place in spite of their shortcomings.” Good, noble reasons. Reasons you might try out when convincing the skeptical or uninitiated.
Then there are the other reasons — the ones that maybe brought you in when you were younger that may be more emotional than high-minded… like “The space battles are awesome!” or the adventure of visiting different planets every week.
Let’s face it, the starships of Star Trek are pretty damned incredible. Able to travel faster than light, protect their crews from the vacuum of space and hostile enemies, able to house a whole community in relative comfort, simulate jazz bars on the holodeck, and make excellent replicated tea on demand. Starships are frickin’ awesome.
There’s one technological marvel aboard ship that probably doesn’t get as much love as it should: sickbay. The engine room has a lot of glamour, science-wise, and the bridge is always where the action is — but dating back to the days of Leonard McCoy, sickbay is also a key part of any interstellar (ahem) enterprise, and it’s no stranger to Star Trek’s spooky ability to predict future technology.
The classic Trek series had its biobeds, and back in the 1960s, a display which combined heart monitor, blood pressure, body temperature, and a few other not-easily-identifiable measures of a patient’s well-being seemed pretty neat — and of course these days, those monitors are now commonplace in a modern hospital.
Beverly Crusher’s sickbay in The Next Generation brought 3D imaging, some very cool heart surgery, live brain simulations, and various therapeutic rays to do things like knit bones or speed up healing in the skin — and the launch of Voyager took it to the next level with the introduction of the Emergency Medical Hologram.
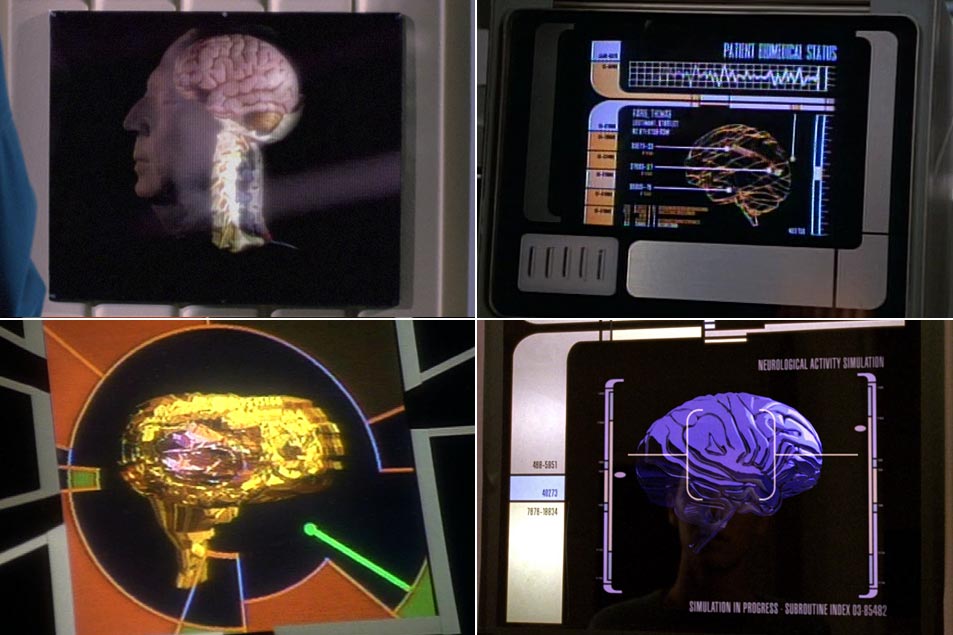
Star Trek has never been shy in speculating about the future of medical technology… even if none of it ever seemed to help Miles O’Brien’s frequently-dislocated shoulder after kayaking in the holosuite.
I’ll admit that it’s a bit of a stretch to claim that dermal regenerators are about to leap off the shelves, but modern research has given us a few things that bring us a bit closer to the medical miracles of the 23rd and 24th centuries — in particular, 3D imaging. MRI or CT scans actually give us a look inside a patient without cutting them open.
While you may not have seen them in your doctor’s office or your local emergency room, there are a few little boxes of tricks that are gaining traction in hospital research labs — ones that as a scientist, I work with every day.
As you might have guessed by all my Star Trek sickbay fanboy stuff, my research is in medical imaging and medical technology. In particular, I’ve spent the last several years working on a thing called diffusion MRI, as part of a whole team in the UK’s National Physical Laboratory at University College London.
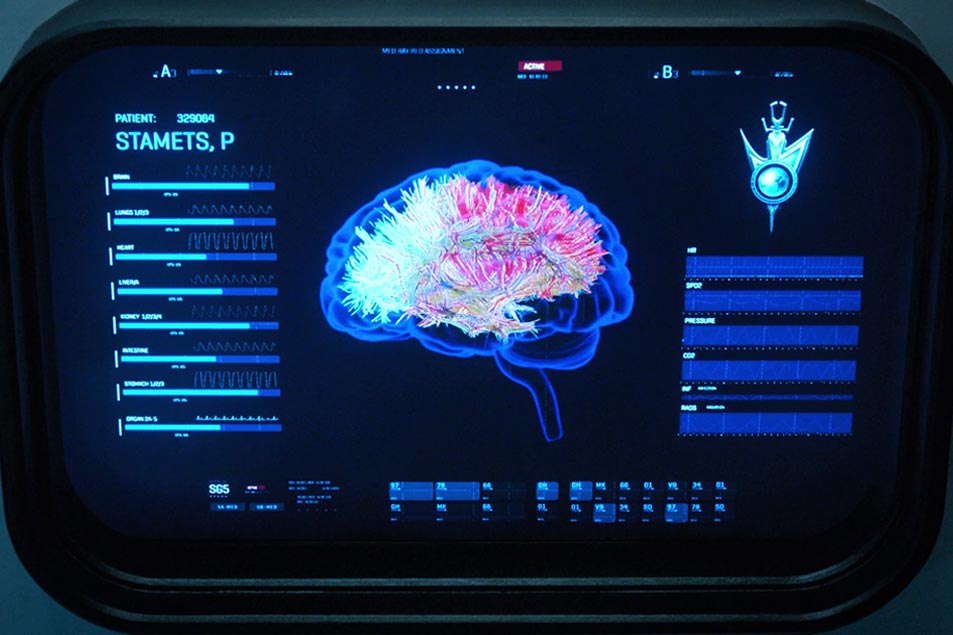
I was excited to see what new medical surprises that Star Trek: Discovery would bring to the franchise, so imagine my delight when diffusion MRI turned up in the new series. Right there on my screen, I watched Sylvia Tilly work with Starfleet technology that’s just nearly right out of my own lab.
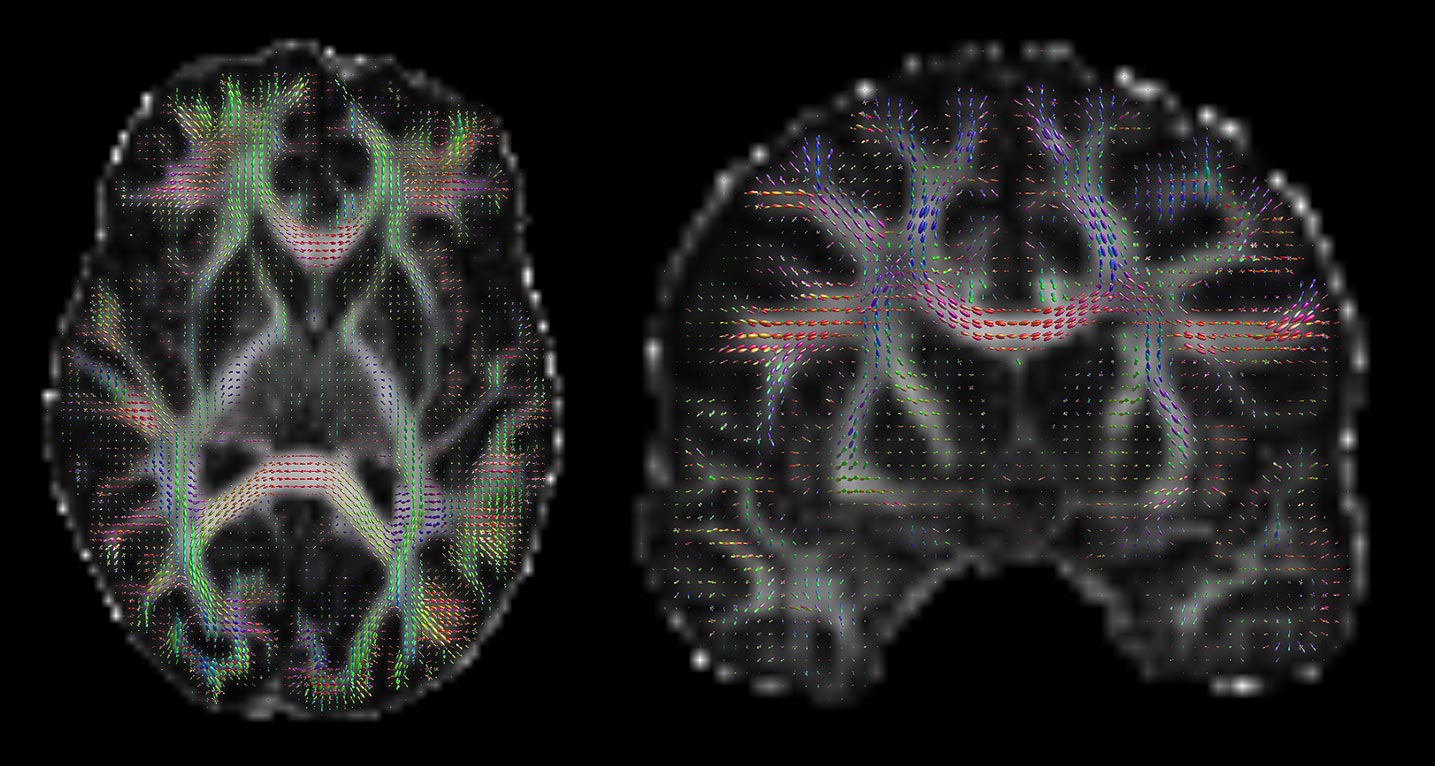
In last season’s “The Wolf Inside,” we see a number of scans of Paul Stamets’ brain — intended to illustrate the damage to his white matter from exposure to the spore drive — and the imagery was clear enough for someone like me to make a decent guess at how the images were created in Discovery’s sickbay.
Here’s the joyous moment: that’s all diffusion MRI imagery! These reconstructions of Stamets’ corpus callosum, a large white matter structure in the brain, are built using a technique called streamline tractography. We use that process in our lab pretty routinely, and even use the images when we work with neurosurgeons.
This is a real thing, that can be done right now, as this video illustrates:
Let’s rewind a bit, though, so I can explain a bit about how one goes about making a 3D map of someone’s white matter. White matter is essentially the ‘cabling’ of your brain, carrying signals from the grey matter and relaying them to places like your muscles when you want to move. (Grey matter is what does your cognitive processing, gets upset, feels happy, receives signals, stores memories, makes decisions, and all that other wonderful stuff your brain gets up to.)
It also carries sensory information and allows the different bits of the brain to exchange information. If your brain was the internet, grey matter is the servers, white matter is the cabling which links them to the peripherals and end users.
Stamets’ brain scan shows the corpus callosum, which is a very large extruded U-shaped tract which allows the two hemispheres of the brain to talk to each other. The close-up is good enough to see that this is constructed with the streamline algorithm I just mentioned, and rendered using straight-line segments. It’s pretty well segmented — in fact, you can see lots of lateral connections, which are easily missed in tractography.

Tilly even states that the scan is showing “changes in gross white matter structure, caused by interactions with the mycelial network,” which is neat, because diffusion and tractography is exactly what you’d use to look for structural abnormality. I have to admit, though, that I haven’t seen a lot of work on diffusion MRI and the mycelial network — but conference season is starting up again, so you never know!
Here, you could compare Stamets’ scan to either a population average, or to earlier scans from his own brain taken before he took on tardigrade duty. It’s all right there — hats off to the show researchers.
Diffusion imaging a particular type of MRI which measures (you guessed it!) diffusion. The human body is full of fluids, and our brains sit in a bath of warm, salty water inside our skulls — and the water is always moving. The molecules jostle and bump into each other randomly and slowly move around in a more-or-less undirected way.
You might remember Brownian motion from high school; that’s the same phenomenon. Dust motes are jostled by the water and slowly spread out, and diffusion imaging allows us to measure how much jostling motion is going on — constructing an image which tells you how much jostling is happening in each pixel. The more jostling, the lower the signal and the darker the pixel.
We can do a lot better than simply ‘dark vs. light,’ though: one of the cool things about the technique is that you can do this in 3D, and get a picture of how things are moving in different directions, in each pixel. From there you can estimate which directions have the most diffusion, and which have the least. There’s a difference between the high school Brownian motion and diffusion in a brain, however: the brain gets in the way.
Diffusing particles don’t just bump into each other, they bump into other things in the tissue, like cell membranes, processing architecture, and cabling — something particularly interesting, because it points in a particular direction.
Imagine being inside a narrow pipe: you can move along the pipe in either direction, but it’s hard to move across it. Flip that around, and if you could measure the direction particles diffuse the furthest, you tell what direction the pipe was pointing. With 50 or 60 different measurements, you can do exactly that.
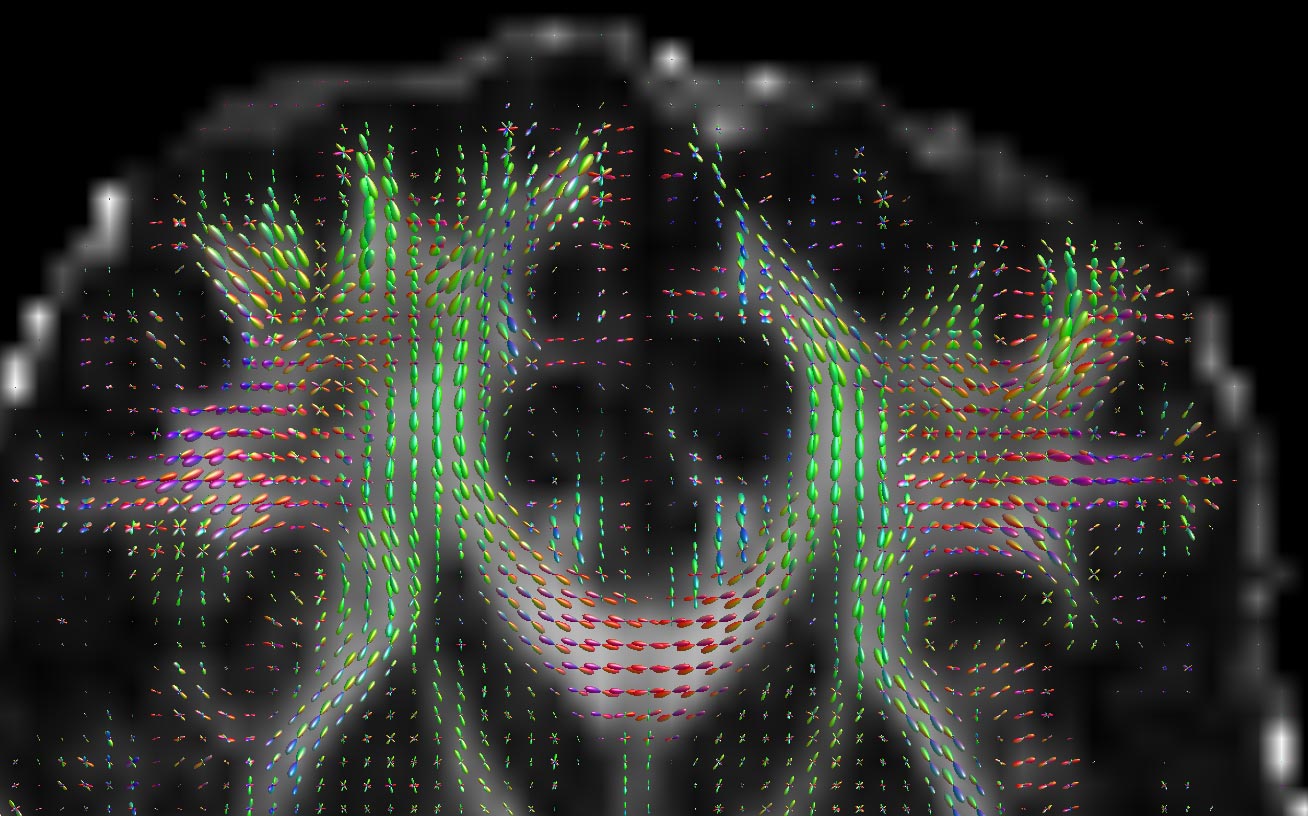
What’s more, you can do it separately in each and every pixel in the image, ending up with images that look a bit like these (courtesy of my friend Donald Tournier’s MRtrix software):
If you look closely, you can see that there’s a little coloured blob in each pixel, which represents the diffusion in 3D. The peaks of the blobs are the directions of most diffusion. We can extract the directions corresponding to the peaks, and we’ve got our estimates of which direction the tissue points.
At this point, we’ve got a load of local orientations; a grid of tiny arrows which tell you which was diffusion is greatest. This works very nicely in cable-like structures like white matter, as we’re estimating local orientations and can even split out the directed tissue from the undirected fairly easily.
We’re not quite at Stamets’ scan yet, though, because there’s still one more thing to do: join those arrows together — this is called tractography. It starts with a 3D field of orientations, and builds up a set of continuous fibres.
There are actually a few different ways to do this, and some are more sophisticated than others, but one standard way is pretty much join-the-dots.
You pick a place to start, and then you follow the field by stepping along the local direction by a small distance, checking the new local orientation, turning in that direction and stepping on. You keep going until there no more directionality — or the structure takes a sharp turn, usually greater than 90 degrees.
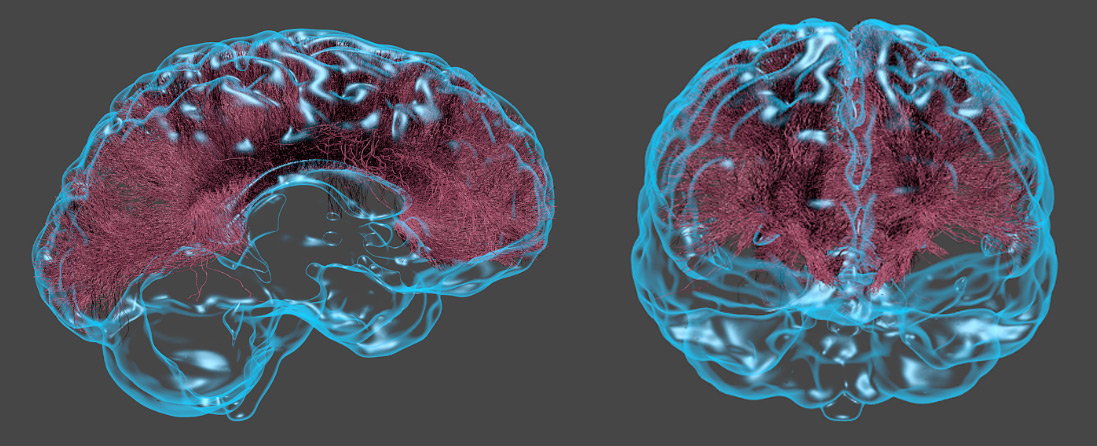
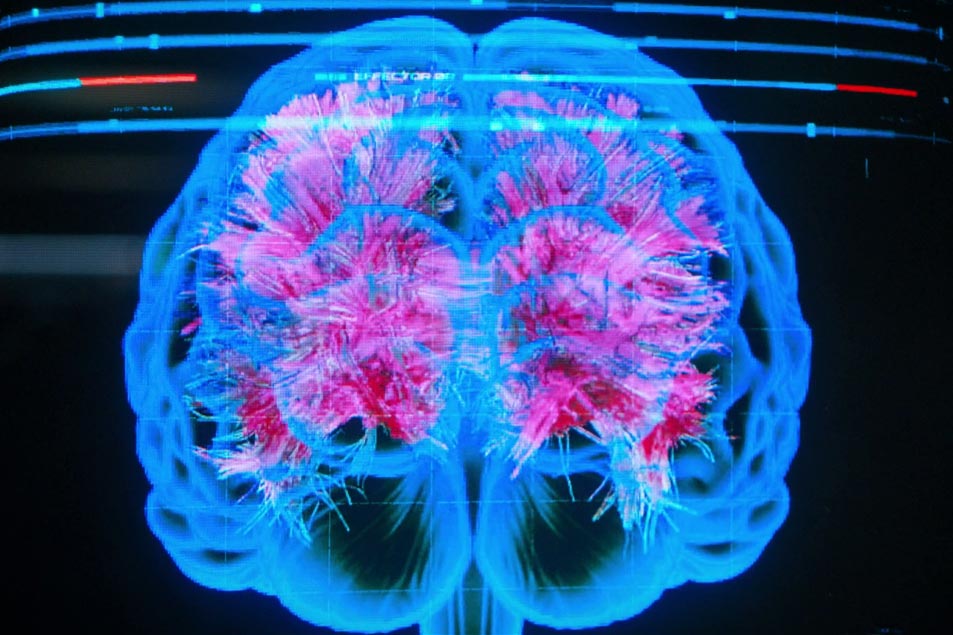
Do this a few thousand times from a bunch of start points, and you can construct what’s called a tract — a big piece of white matter which looks like the sort of thing you’d find in a medical textbook — and with that in mind, we reach our lab’s version of Stamets’ brain scan:
This is a reconstruction of the corpus callosum using diffusion MRI and streamline tractography. It’s the same data from before, but with the streamlines built up. We’ve also added a 3D cortical surface from the same scan… which is there mostly to make it look cool. This isn’t Stamets’ brain, of course; it’s actually a scan of Kiran Seunarine, our resident tractography expert. (To the best of my knowledge, he’s not able to navigate the mycelial network yet…)
So if we’re not scanning for spore drive brain issues, why might you want to do this in real life? Well, one of the nice things about tractography is that it shows you the 3D structure in each individual patient, not just a textbook version of a brain. This might be interesting if you’re studying how brains change during aging or natural development, for example, but it’s also useful if something goes wrong: brain tumours, for example.
A growing tumour pushes the tissue around it aside or can grow right over the top of something else. If a surgeon is operating to remove it, the biggest risks are either not removing all or it — in which case it could grow back — or removing too much, taking some healthy tissue out as well.
There’s also the question of how you get to the thing in the first place. You want to avoid cutting through white matter, because once it’s gone, it’s gone — and a broken cable in the brain is like a broken cable in any other network. Information can’t get through, except in this case that blocked sensory information might be coming from your eyes or fingertips. You want that information to get through.

One of the things we do in our lab is tractography for neurosurgeons, used in reconstructing white matter around, say, a tumour, so the surgeons can use it to plan the procedure. The more information they can get, the better they can plan, and the better patients will recover.
Tilly also uses imaging to monitor how well a treatment is working, and we do that too. We also use diffusion imaging and tractography to look at brain maturation — from early childhood to adolescence — to learn more about what’s happening in healthy kids, and when they’re suffering from a disease like epilepsy or multiple sclerosis. There’s a lot you can tell from white matter.
Diffusion MRI technology is a little bit of 23rd century sickbay magic that’s actually already here. As far as I know, no one is using it to scan tardigrade brains quite yet, but I’m trying to see if my colleagues in the lab will let me render everything in Discovery’s pink and blue color scheme to bring that Starfleet feeling back down to Earth.
![]()
Matt Hall is a research scientist at University College London and the UK’s National Physical Laboratory working with advanced MRI technology to develop the tools needed to obtain the most detailed and accurate images possible of human tissue. He is part of a team of scientists and clinicians at The UCL Institute of Child Health who develop and apply new MRI methods, particularly for childhood illnesses.
The images in this article were created by Kiran Seunarine, a scientist with a background in design and visualisation — a modern-day Geordi La Forge who works with magnets instead of warp reactors.